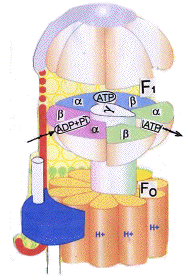
structure, which, in the case of mitochondria, sticks out into the matrix and is anchored to the
membrane by a stator to prevent rotation. It consists of three α- and three β- subunits, all of which can bind nucleotides, but
only the β-subunits can take part in the reactions. Fo is a cylindrical structure capable of rotation when driven by translocated
protons, and which is linked to a central stalk that can revolve inside F1.
The mechanism that drives ATP synthesis seems to depend upon a binding charge conception in which catalytic sites on the β-subunits
have different affinities for nucleotides and are designated loose (L), tight (T), and
Open(O). .These are coloured pink, blue, and
green respectively. The loose (L) sites bind the substrates (ADP and phosphate)
|
 Click on the image to
view the animation. Click on the image to
view the animation.
|
O, and O to L
(after releasing ATP). The new L site then binds new ADP and phosphate and begins
a new reaction sequence. One complete revolution of Fo therefore results in the formation of 3ATP, one from each of the β-subunits.
In this example about 10 protons need to be retrolocated for each complete revolution of Fo, which means that the formation of 1ATP
requires about 3.3, though other species may be different. ATP synthase is thought to revolve at more than 100Hz (revolutions/sec.),
which is sufficient to produce a turnover of the equivalent of the weight of our body of ATP each day.
Many other subunits are involved, some of which are understood, but others are the centre of on-going research. This
animation is a
simplified "interpretation" of the complex integrated movements which
|